2 Creating Reproducible Documents
In this chapter, we introduce you to using code to create reproducible research. Creating reproducible research means you will write text and code that completely and transparently performs an analysis from start to finish in a way that produces the same result for different people using the same software on different computers. We will cover things such as file structure and setting a working directory, using R Markdown files, and writing code chunks.
As well as improving transparency with others researchers, reproducible research benefits you. When you return to an analysis or task after days, weeks, or months, you will thank past you for doing things in a transparent, reproducible way, as you can easily pick up right where you left off.
Chapter Intended Learning Outcomes (ILOs)
By the end of this chapter, you will be able to:
Understand and set your working directory, either manually or through creating an R project.
Create a knit an R Markdown file to create a reproducible document.
Use inline code to combine text and code output in your reproducible documents.
Identify and fix common errors in knitting R Markdown files.
2.1 File structure, working directories, and R Projects
In chapter 1, we never worked with files, so you did not have to worry about where you put things on your computer. Before we can start working with R Markdown files, we must explain what a working directory is and how your computer knows where to find things.
Your working directory is the folder where your computer starts to look for files. It would be able to access files from within that folder and within sub-folders in your working directory, but it would not be able to access folders outside your working directory.
In this course, we are going to prescribe a way of working to support an organised file system, helping you to know where everything is and where R will try to save things on your computer and where it will try to save and load things. Once you become more comfortable working with files, you can work in a different way that makes sense to you, but we recommend following our instructions for at least RM1 as the first course.
2.1.1 Activity 1: Create a folder for all your work
In your documents or OneDrive, create a new folder called ResearchMethods1_2. This will be your highest level folder where you will save everything for Research Methods 1 and 2.
When you are a student at the University of Glasgow, you have access to the full Microsoft suite of software. One of those is the cloud storage system OneDrive. We heavily recommend using this to save all your work in as it backs up your work online and you can access it from multiple devices.
Within that folder, create two new folders called Assessments and Quant_Fundamentals. In Assessments, you can save all your assessments for RM1 and RM2 as you come to them. In Quant_Fundamentals, that is where you will save all your work as you progress through this book.
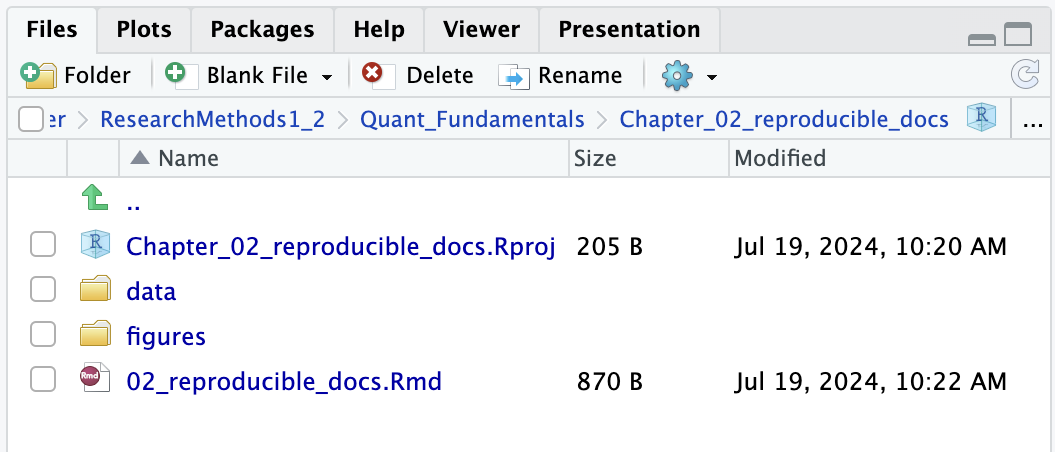
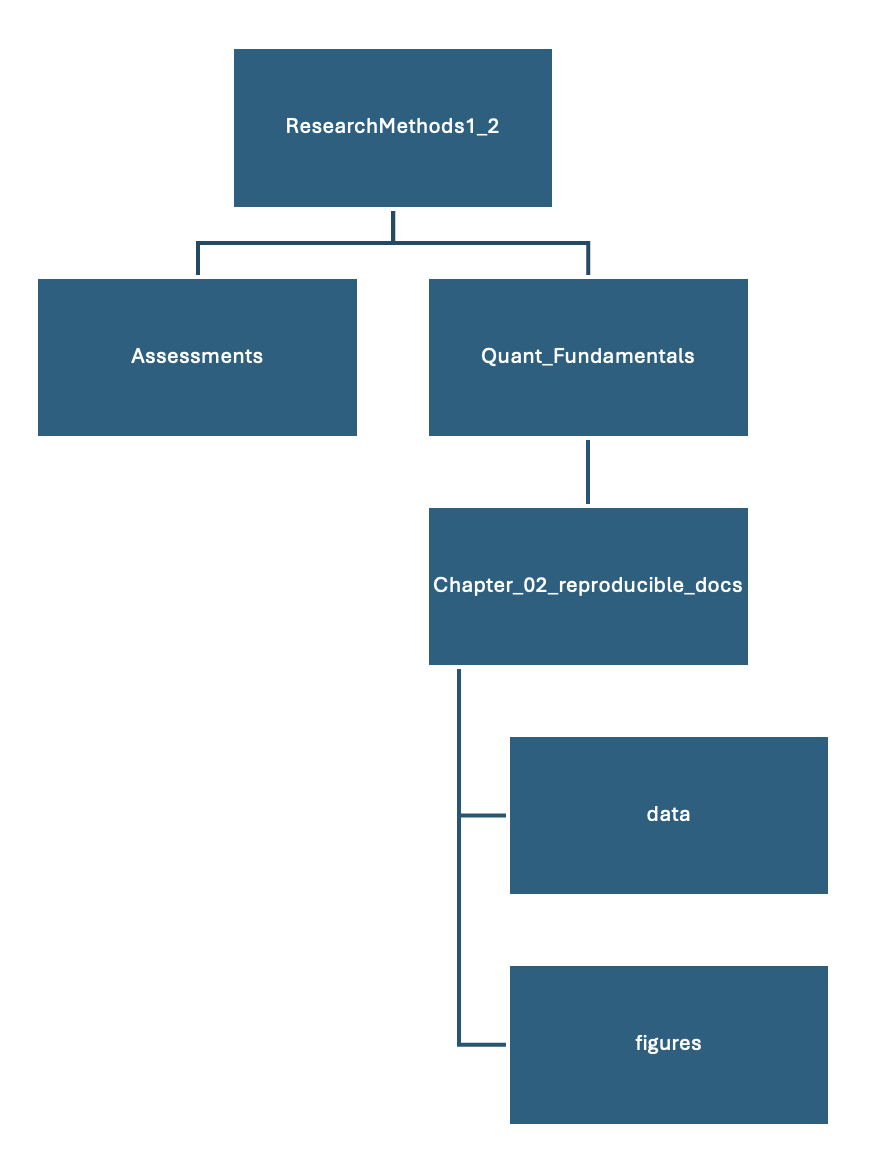
Within Quant_Fundamentals, create a new folder called Chapter_02_reproducible_docs. As you work through the book, you will create a new chapter folder each time you start a new chapter and the sub-folders will always be the same. Within Chapter_02_reproducible_docs, create two new folders called data and figures. As a diagram, it should look like Figure 2.1.
You might notice in the folder names we avoided using spaces by adding things like underscores _ or capitalising different words. Historically, spaces in folder/file names could cause problems for code, but now it’s just slightly easier when file names and folder names do not have spaces in them.
For naming files and folders, try and choose something sensible so you know what it refers to. You are trying to balance being as short as possible, while still being immediately identifiable. For example, instead of fundamentals of quantitative analysis, we called it Quant_Fundamentals.
When you create and name folders to use with R / RStudio, whatever you do, do not call the folder “R”. If you do this, sometimes R has an identity crisis and will not save or load your files properly. It can also really damage your setup and require you to reinstall everything as R tends to save all the packages in a folder called R. If there is another folder called R, then it gets confused and stops working properly.
If we support you to use the online University of Glasgow R Server, working with files is a little different. If you downloaded R / RStudio to your own computer or you are using one of the library/lab computers, please ignore this section.
The main disadvantage to using the R server is that you will need create folders on the server and then upload and download any files you are working on to and from the server. Please be aware that there is no link between your computer and the R server. If you change files on the server, they will not appear on your computer until you download them from the server, and you need to be very careful when you submit your assessment files that you are submitting the right file. This is the main reason we recommend installing R / RStudio on your computer wherever possible.
Going forward throughout this book, if you are using the server, you will need to follow an extra step where you also upload them to the sever. As an example:
Log on to the R server using the link we provided to you.
In the file pane, click
New folderand create the same structure we demonstrated above.Download
ahi-cesd.csvandparticipant-info.csvinto thedatafolder you created for chapter 2. To download a file from this book, right click the link and select “save link as”. Make sure that both files are saved as “.csv”. Do not open them on your machine as often other software like Excel can change setting and ruin the files.Now that the files are stored on your computer, go to RStudio on the server and click
UploadthenBrowseand choose the folder for the chapter you are working on.Click
Choose fileand go and find the data you want to upload.
2.1.2 Manually setting the working directory
Now that you have a folder structure that will keep everything nice and organised, we will demonstrate how you can manually set the working directory. If you open RStudio, you can check where the current working directory is by typing the function getwd() into the console and pressing enter/return. That will show you the current file path R is using to navigate files. If you look at the Files window in the bottom right, this will also show you the files and folders available from your working directory.
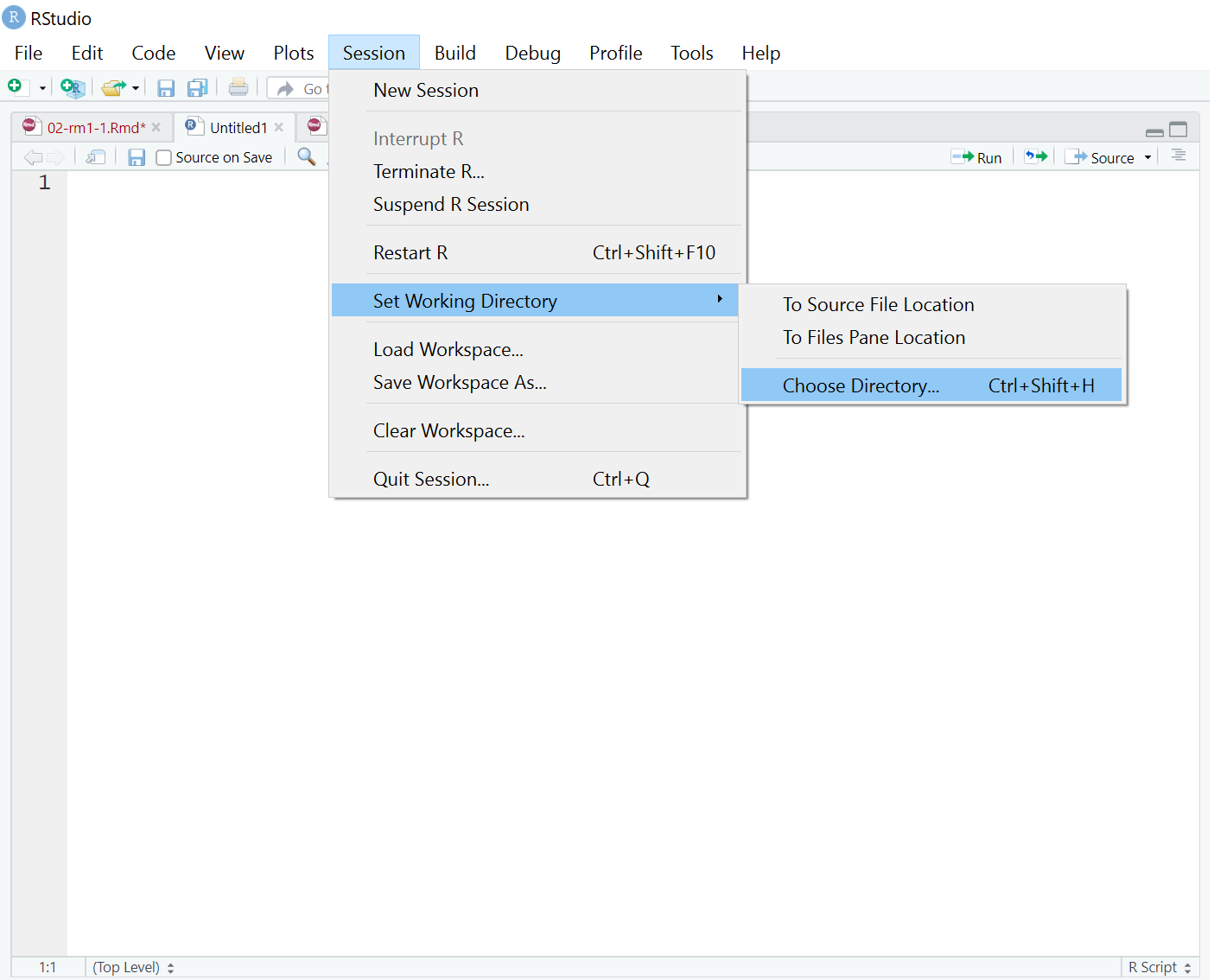
If you click on the top menu Session >> Set Working Directory >> Choose Directory..., (Figure 2.2) you can navigate through your documents or OneDrive until you can select Chapter_02_reproducible_docs. Click open and that will set the folder as your working directory. You can double check this worked by running getwd() again in the console.
2.1.3 Activity 2 - Creating an R Project
Knowing how to check and manually set your working directory is useful, but there is a more efficient way of setting your working directory alongside organised file management. You are going to create something called an R Project.
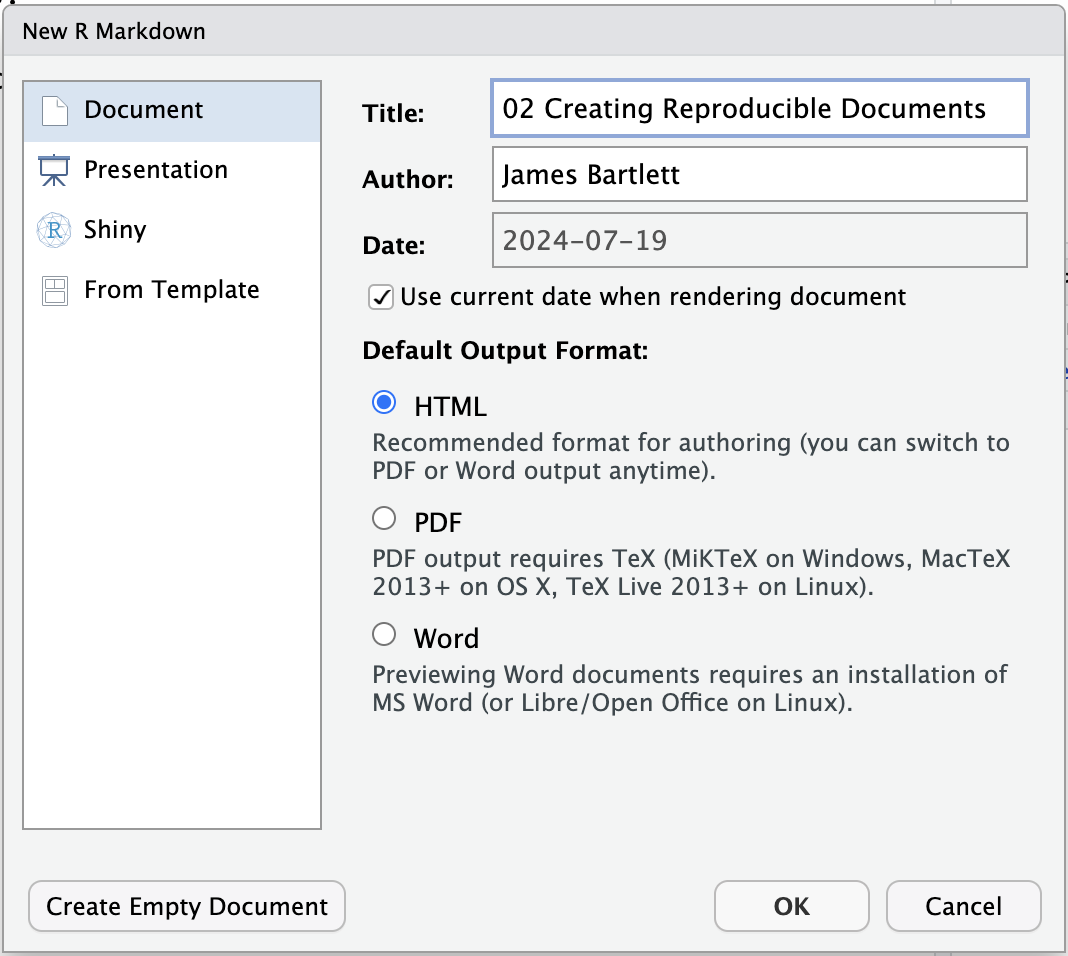
To create a new project for the work you will do in this chapter (Figure 2.3):
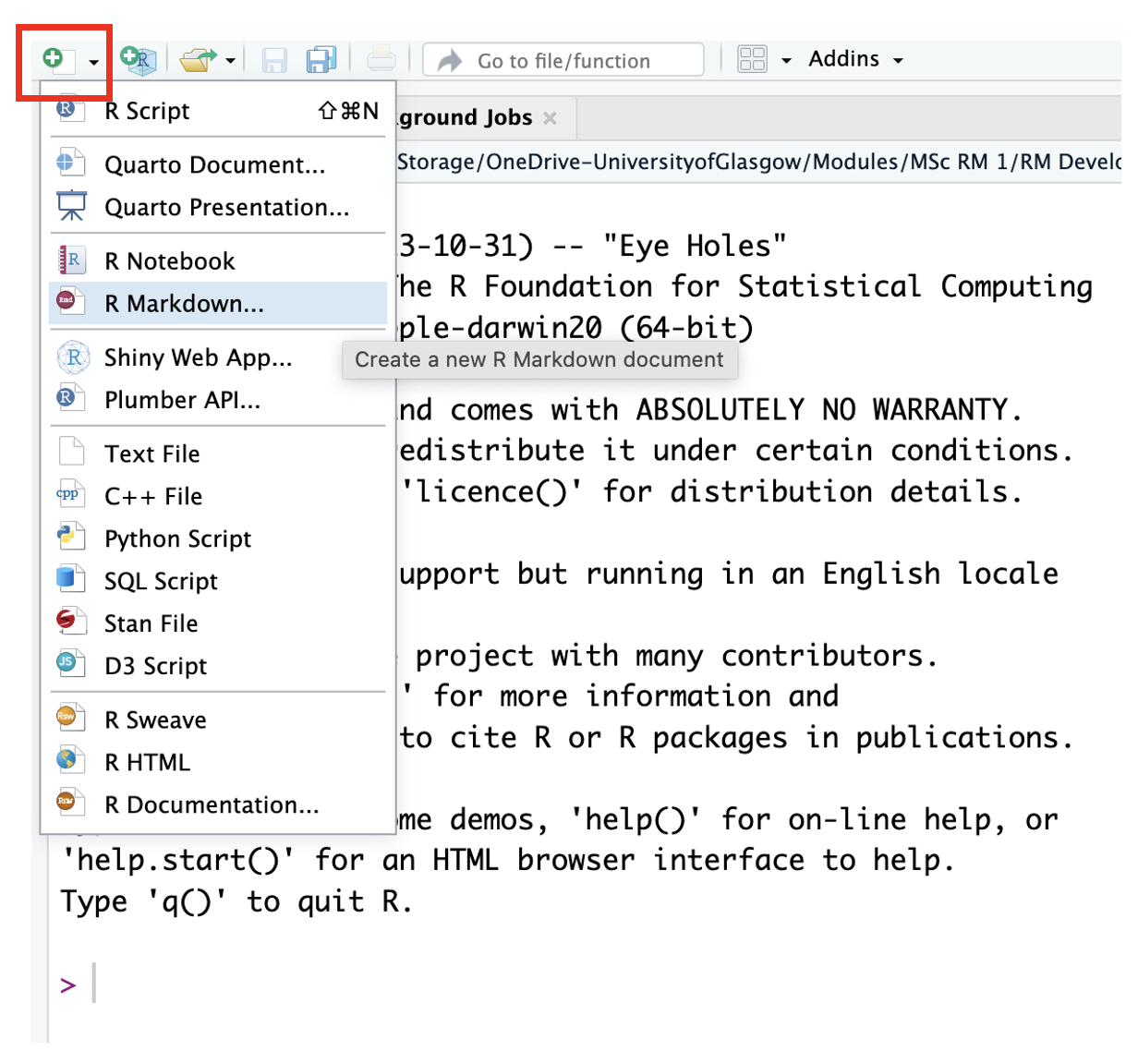
Click on the top menu and navigate to
File >> New Project....You have the option to select from New Directory, Existing Directory, or Version Control. You already created a folder for
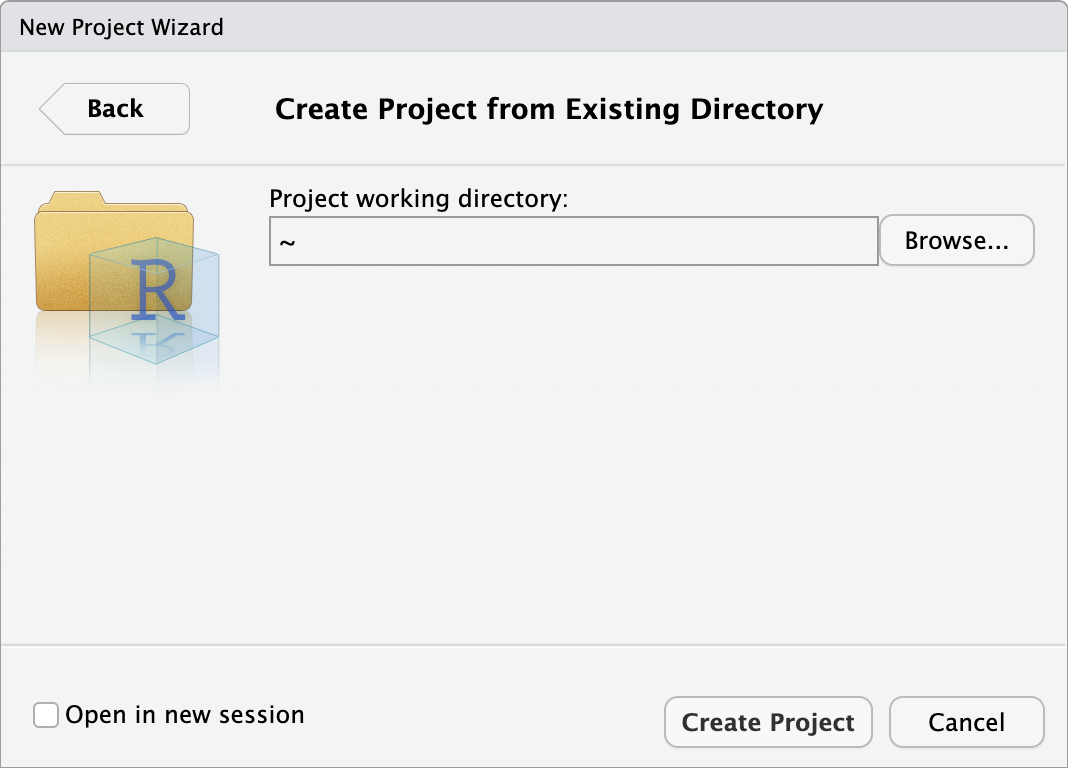
Chapter_02_reproducible_docs, so select Existing Directory.Click Browse… next to Project working directory to select the folder you want to create the project in.
When you have navigated to
Chapter_02_reproducible_docsfor this chapter, click Open and then Create Project.
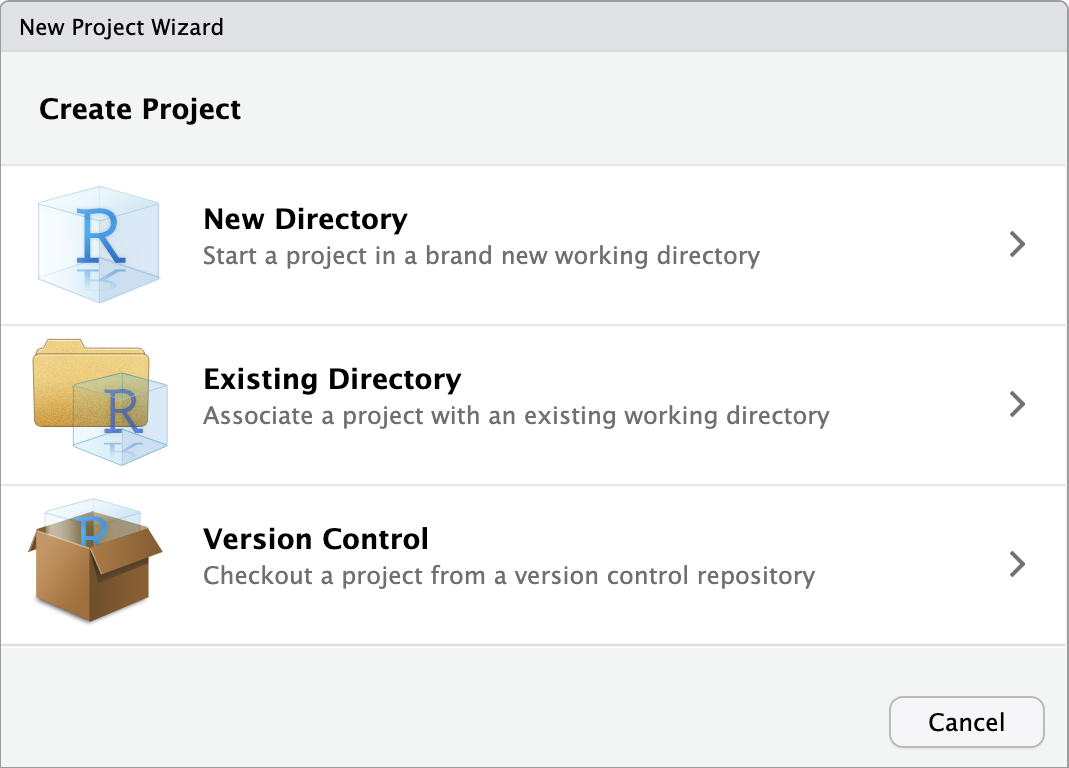
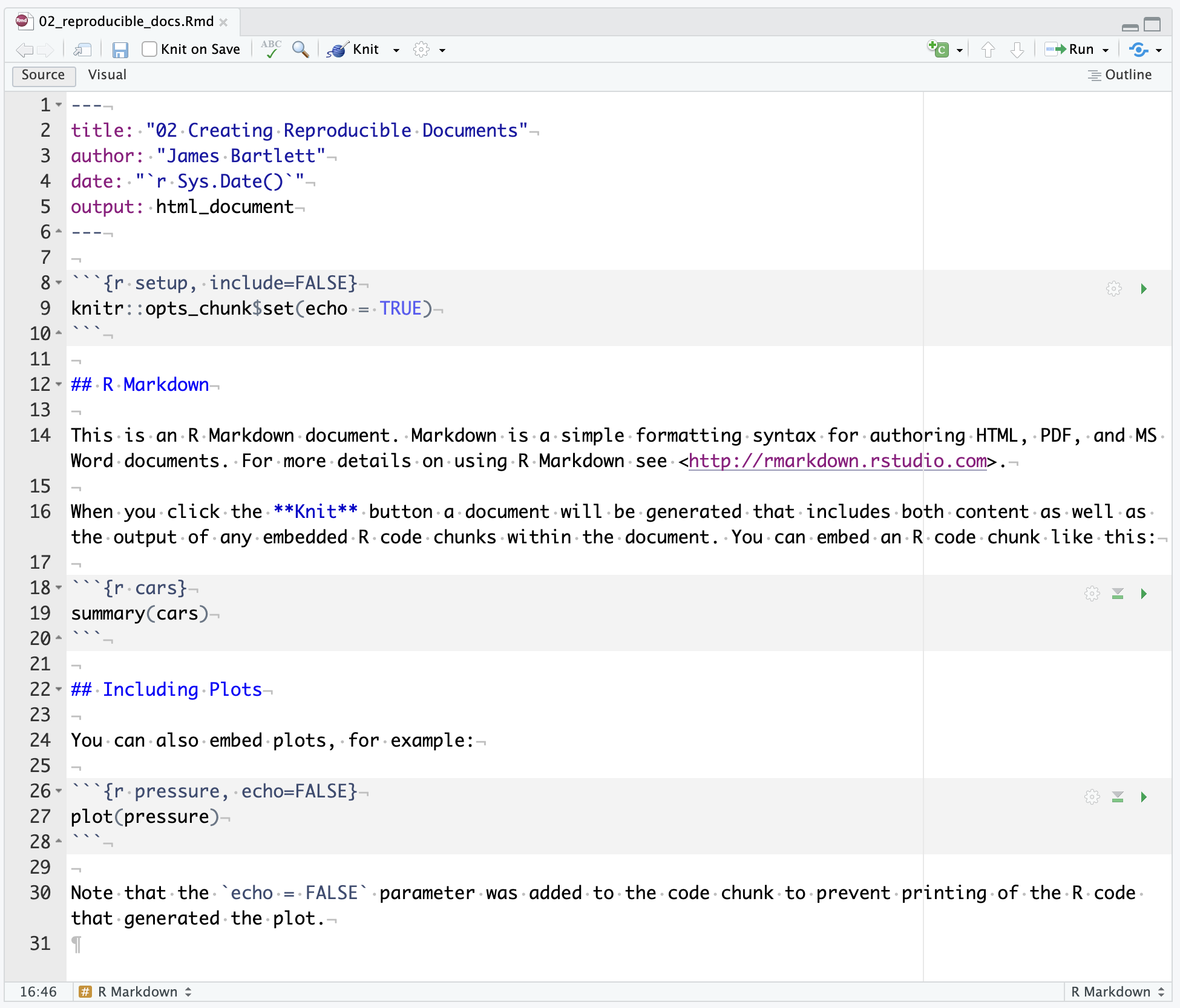
RStudio will restart itself and open with this new project directory as the working directory. You should see something like Figure 2.4.
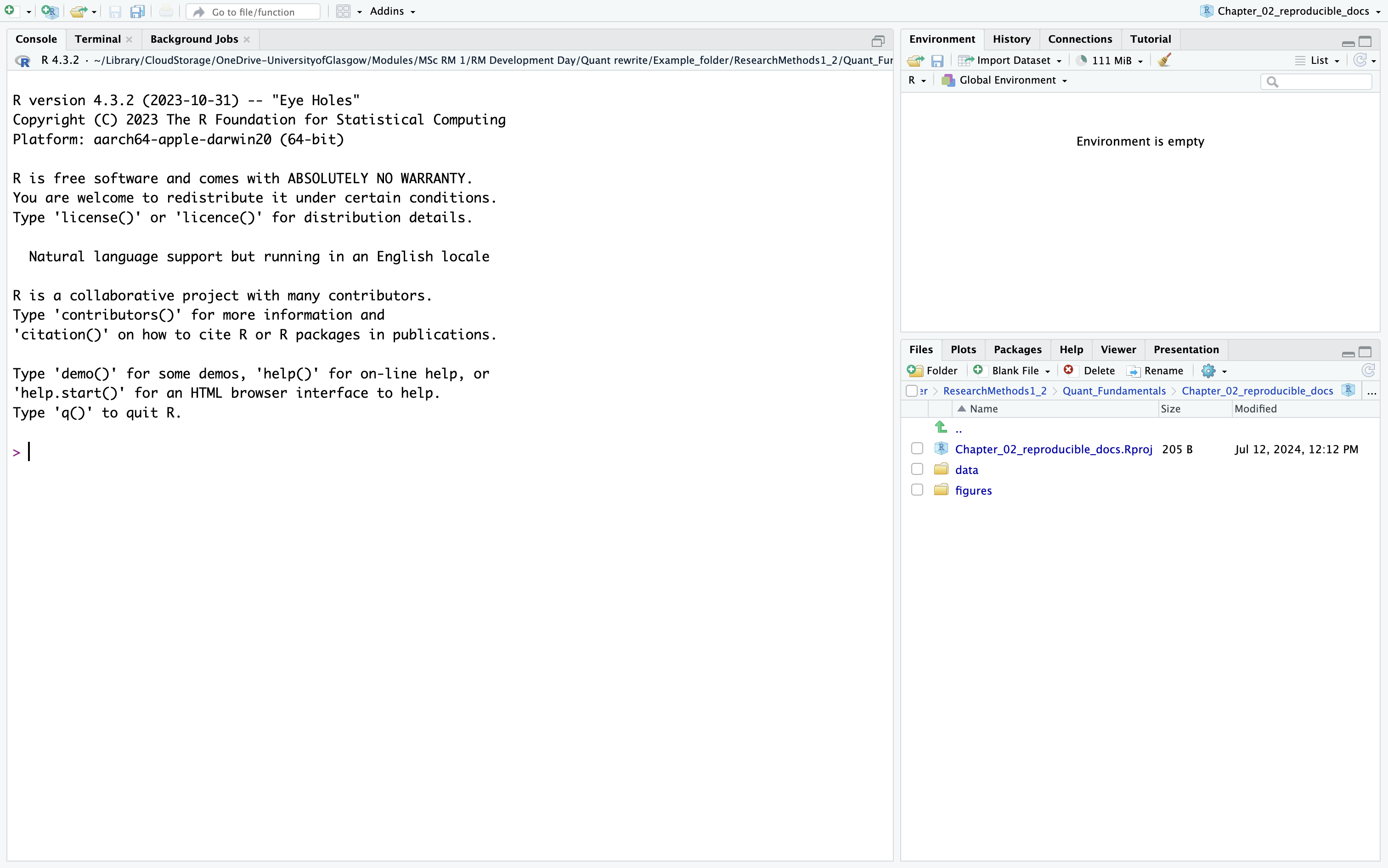
In the files tab in the bottom right window, you will see all the contents in your project directory. You can see your two sub-folders for data and figures and a file called Chapter_02_reproducible_docs.Rproj. This is a file that contains all of the project information. When you come back to this project after closing down RStudio, if you double click on the .Rproj file, it will open up your project and have your working directory all set up and ready to go.
In each chapter, we will repeat these instructions at the start to prescribe this file structure, but when you create your own folders and projects, do not ever save a new project inside another project. This can cause some hard to resolve problems. For example, it would be fine to create a new project within the Quant_Fundamentals folder as we will do for each new chapter, but should never create a new project within the Chapter_02_reproducible_docs folder.
2.3 Demonstrating reproducibility
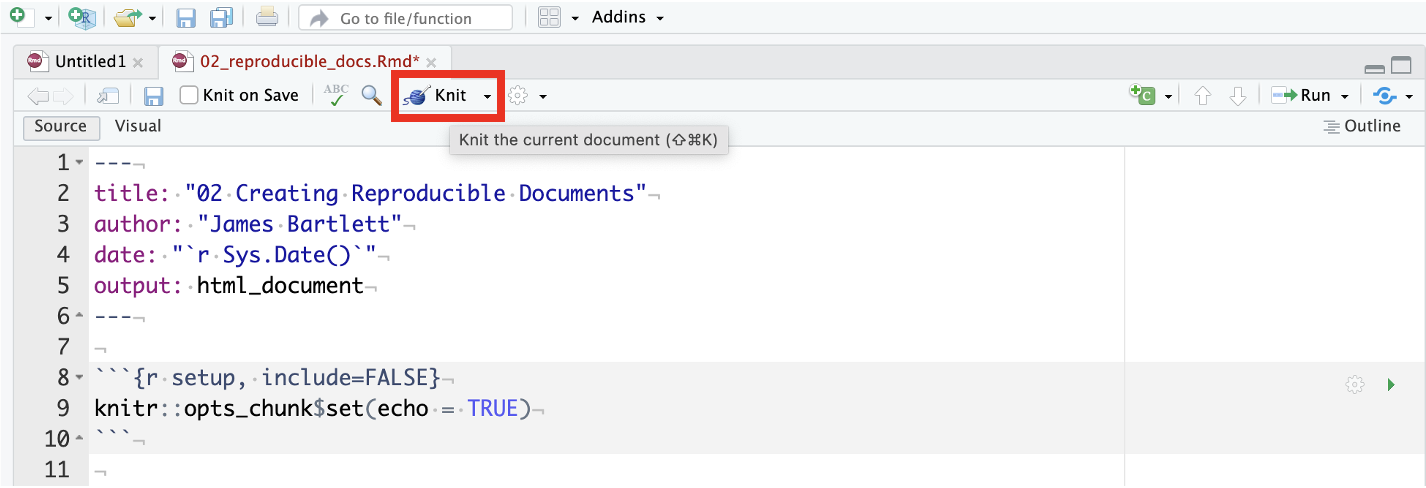
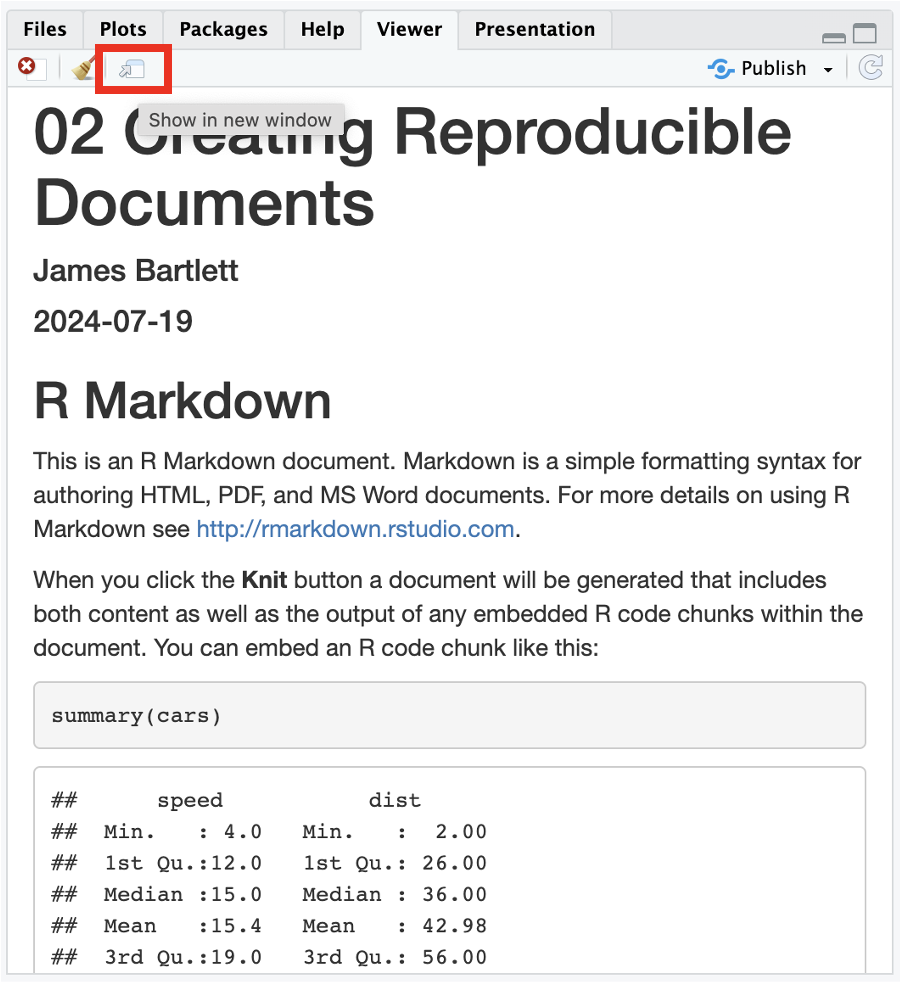
At the start of this chapter, we plugged the benefits of reproducible research as the ability to produce the same result for different people using the same software on different computers. We are going to end the chapter on a demonstration of this by giving you an R Markdown document and data. You should be able to click knit and see the results without editing anything. We do not expect you to understand the code included in it, we are previewing the skills you will develop over the next four chapters on visualisation and data wrangling.
2.3.1 Activity 9 - Knit the reproducibility demonstration document
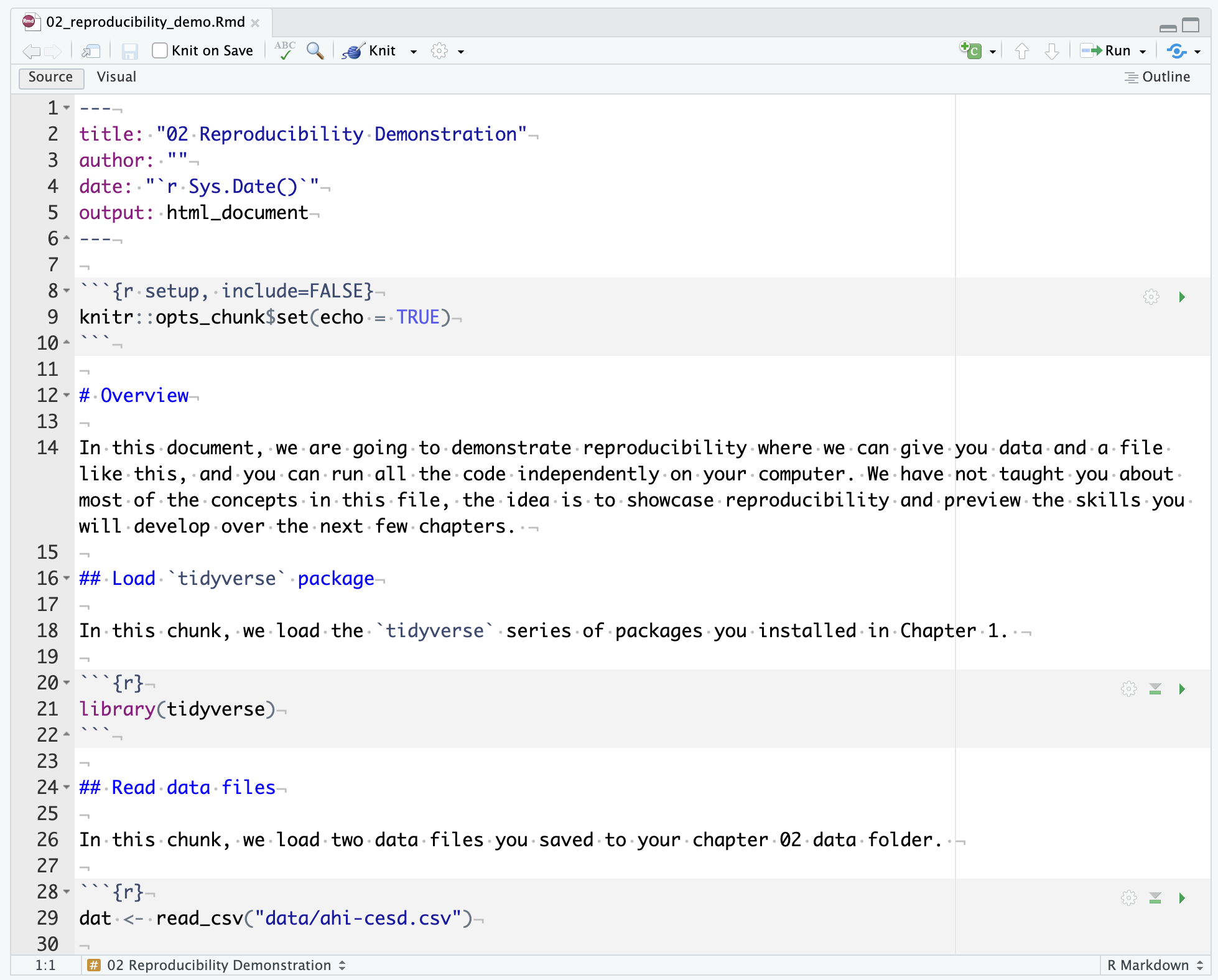
Please follow these steps and you should be able to knit the document without editing anything. Make sure you are still in your Chapter_02_reproducible_docs folder. If you are coming back to this activity, remember to set your working directory by opening the .Rproj file.
If you are working on your own computer, make sure you installed the
tidyversepackage. Please refer to Chapter 1 - Activity 3 if you have not completed this step yet. If you are working on a university computer or the online server, you do not need to complete this step astidyversewill already be installed.Download the R Markdown document through the following link: 02_reproducibility_demo.Rmd. To download a file from this book, right click the link and select “save link as”, or just clicking the link will save the file to your Downloads. Save or copy the file to your
Chapter_02_reproducible_docsfolder.Download these two data files. Data file one: ahi-cesd.csv. Data file two: participant-info.csv. Right click the links and select “save link as”, or clicking the links will save the files to your Downloads. Make sure that both files are saved as “.csv”. Do not open them on your machine as often other software like Excel can change setting and ruin the files. Save or copy the file to your
data/folder withinChapter_02_reproducible_docs.
At this point, you should have “02_reproducibility_demo.Rmd” within your Chapter_02_reproducible_docs folder. You should have “ahi-cesd.csv” and “participant_info.csv” in the data/ folder within Chapter_02_reproducible_docs.
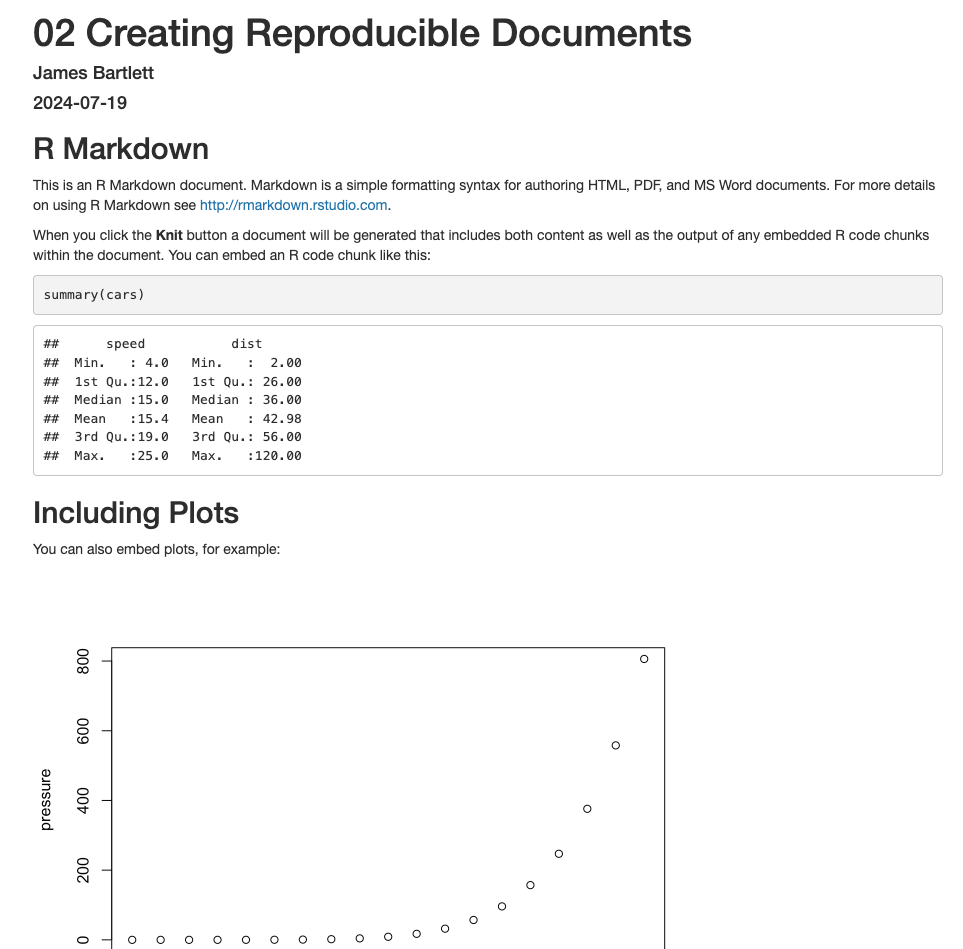
If you open “02_reproducibility_demo.Rmd” and followed all the steps above, you should be able to click knit. This will turn the R Markdown file into a knitted html file, showing some data wrangling, summary statistics, and two graphs (Figure 2.14). In the next chapter, you will learn how to write this code yourself, starting with creating graphs.
If you have any questions or problems about anything contained in this chapter, please remember you are always welcome to post on the course Teams channel, attend a GTA support session, or attend the office hours of one of the team.
2.4 Test yourself
To end the chapter, we have some knowledge check questions to test your understanding of the concepts we covered in the chapter. We then have some error mode tasks to see if you can find the solution to some common errors in the concepts we covered in this chapter.
2.4.1 Knowledge check
Question 1. One of the key first steps when we open RStudio is to:One of the most common issues we see where code does not work the first time is because people have forgotten to set the working directory. The working directory is the starting folder on your computer where you want to save any files, any output, or contains your data. R/RStudio needs to know where you want it to look, so you must either manually set your working directory, or open a .Rproj file.
Question 2. When using the default environment color settings for RStudio, what color would the background of a code chunk be in R Markdown?
Question 3. When using the default environment color settings for RStudio, what color would the background of normal text be in R Markdown?
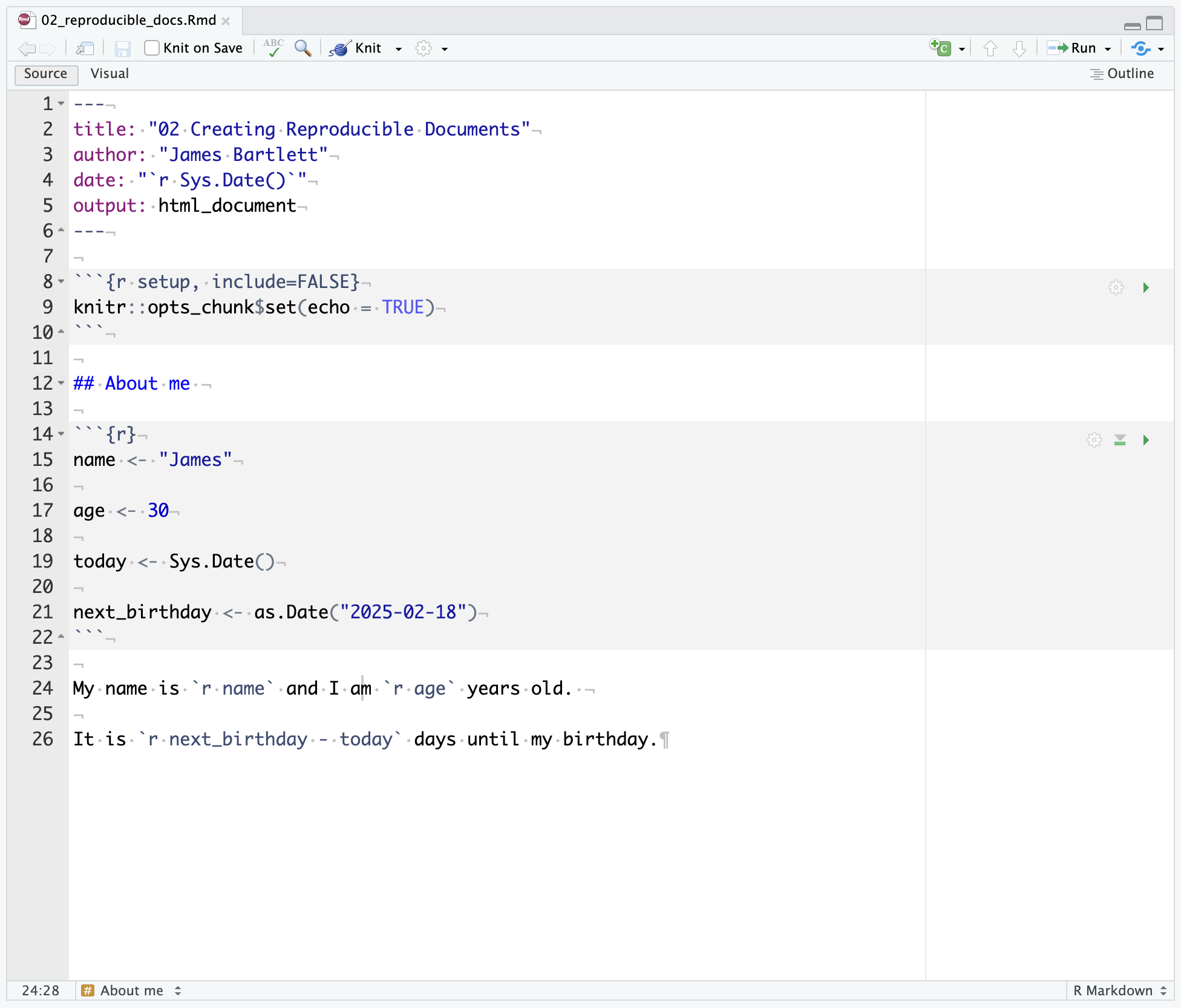
Assuming you have not changed any of the settings in RStudio, code chunks will tend to have a grey background and normal text will tend to have a white background. This is a good way to check that you have closed and opened code chunks correctly.
Code chunks always take the same general format of three backticks followed by curly parentheses and a lower case r inside the parentheses ({r}). People often mistake these backticks for single quotes but that will not work. If you have set your code chunk correctly using backticks, the background color should change to grey from white
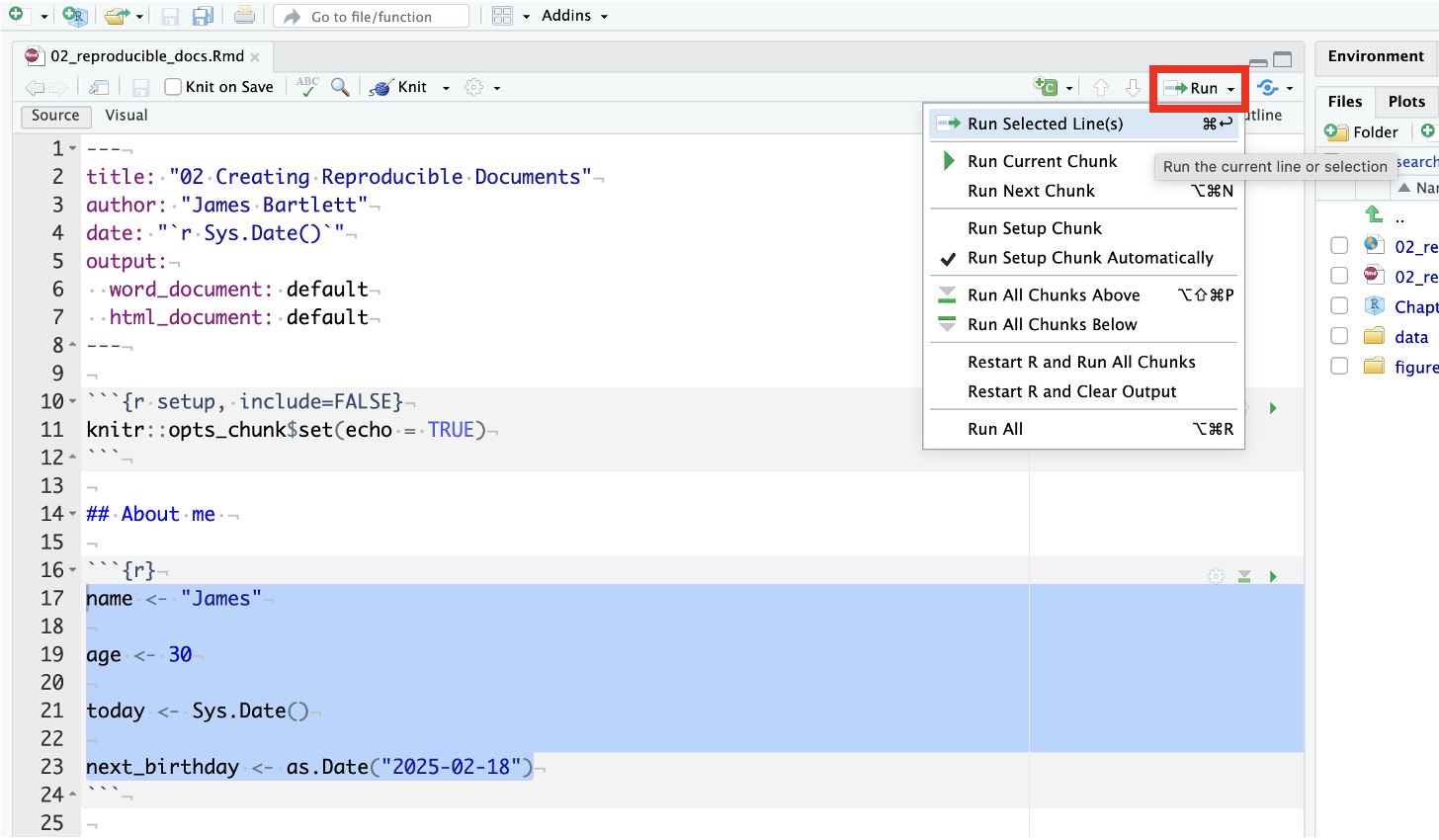
Inline coding is an incredibly useful approach for merging text and code in a sentence outside of a code chunk. It can be really useful for when you want to add values from your code directly into your text. If you copy and paste values, you can easily create errors, so it’s useful to add inline code where possible.
2.4.2 Error mode
The following questions are designed to introduce you to making and fixing errors. For this topic, we focus on R Markdown and potential errors in using code blocks and inline code. Remember to keep a note of what kind of error messages you receive and how you fixed them, so you have a bank of solutions when you tackle errors independently.
Create and save a new R Markdown file by following the instructions in activity 3 and activity 4. You should have a blank R Markdown file below line 10. Below, we have several variations of a code chunk and inline code errors. Copy and paste them into your R Markdown file, click knit, and look at the error message you receive. See if you can fix the error and get it working before checking the answer.
Question 5. Copy the following text/code/code chunk into your R Markdown file and press knit. You should receive an error like Error while opening file. No such file or directory.
Here, we wrote city <- "Glasgow" outside the code chunk. So, when we try and knit, it is not evaluated as code, and city does not exist as an object to be referenced in inline code. If you copy city <- "Glasgow" into the code chunk and press knit, it should work.
Question 6. Copy the following text/code/code chunk into your R Markdown file and press knit. You should receive an error like Error in parse(): ! attempt to use zero-length variable name which is not very helpful for diagnosing the problem.
Here, we missed a final backtick in the code chunk. You might have noticed all the text had a grey background, so R Markdown thought everything was code. So, when it reached the inline code and text, it tried interpreting it as code and caused the error. If you add the final backtick to the code chunk, you should be able to click knit successfully.
Question 7. Copy the following text/code/code chunk into your R Markdown file and press knit. You should receive an error like Error while opening file. No such file or directory.
Here, we tried using inline code before the code chunk. R Markdown runs the code from start to finish in a fresh environment. We tried referencing city in inline code, but R Markdown did not know it existed yet. To fix it, you need to move the inline code below the code chunk, so you create city before referencing it in inline code.
Question 8. Copy the following text/code/code chunk into your R Markdown file and press knit. This…works?
Here, we have a sneaky kind of “error” where it knits, but it is not doing what we wanted it to do. In the inline code part, we only added code formatting city, we did not add the r to get R Markdown to interpret it as R code:
If you add the r after the first backtick, it should knit and add the city object in.
2.5 Words from this Chapter
Below, you will find a list of words that we used in this chapter that might be new to you in case you need to refer back to what they mean. The links in this table take you to the entry for the words in the PsyTeachR Glossary. Note that numerous members of the team wrote entries in the Glossary and as such the entries may use slightly different terminology from what we used in the chapter.
| term | definition |
|---|---|
| chunk | A section of code in an R Markdown file |
| html | Hyper-Text Markup Language: A system for semantically tagging structure and information on web pages. |
| inline-code | Directly inserting the result of code into the text of a .Rmd file. |
| knit | To create an HTML, PDF, or Word document from an R Markdown (Rmd) document |
| latex | A typesetting program needed to create PDF files from R Markdown documents. |
| markdown | A way to specify formatting, such as headers, paragraphs, lists, bolding, and links. |
| r-markdown | The R-specific version of markdown: a way to specify formatting, such as headers, paragraphs, lists, bolding, and links, as well as code blocks and inline code. |
| r-project | A project is simply a working directory designated with a .RProj file. When you open an R project, it automatically sets the working directory to the folder the project is located in. |
| reproducible-research | Research that documents all of the steps between raw data and results in a way that can be verified. |
| working-directory | The filepath where R is currently loading files from and saving files to. |
2.6 End of chapter
Well done on reaching the end of the second chapter! This was another long chapter as we had to cover a range of foundational skills to prepare you for learning more of the coding element in future chapters.
The next chapter builds on all the skills you have developed so far in R programming and creating reproducible documents to focus on something more tangible: data visualisation in R to create plots of your data.